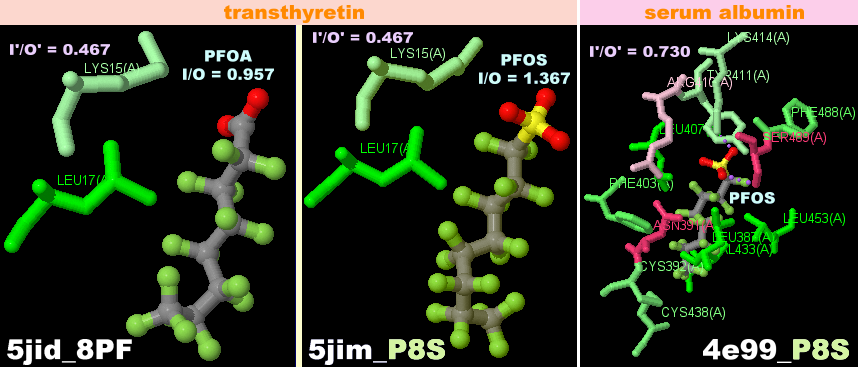
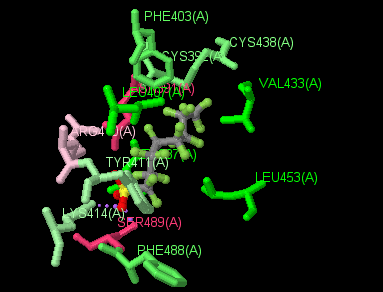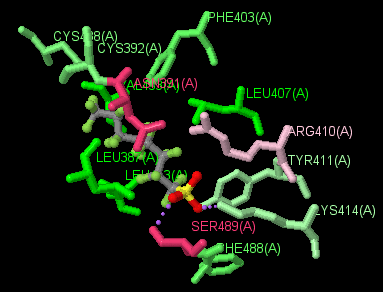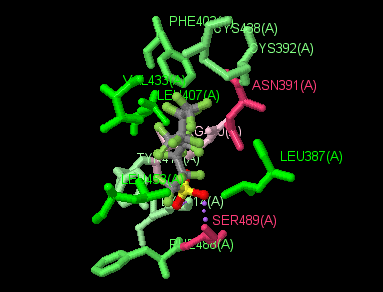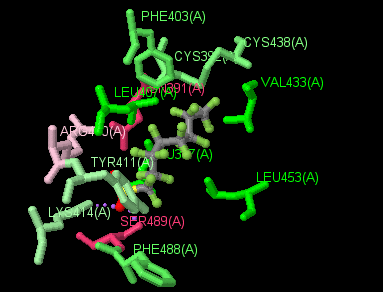
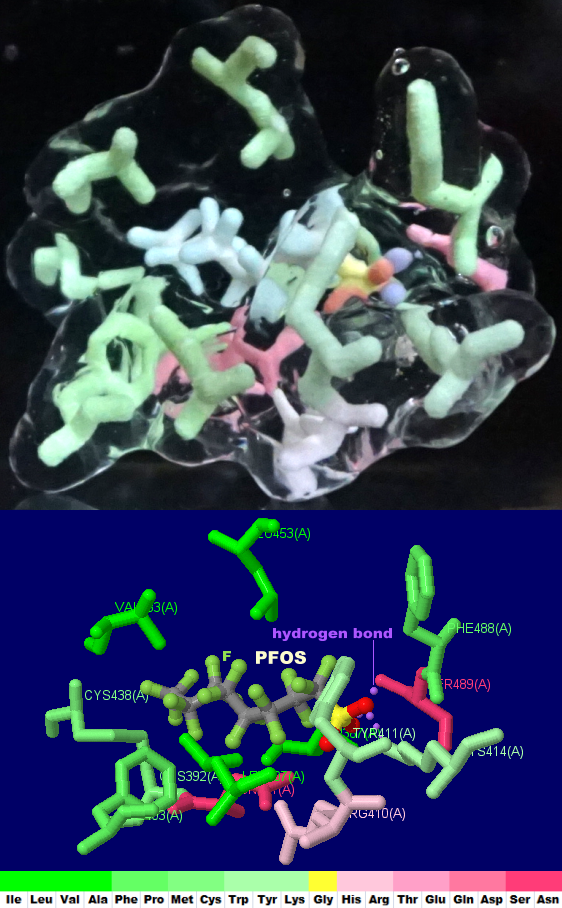
(Jmol version)
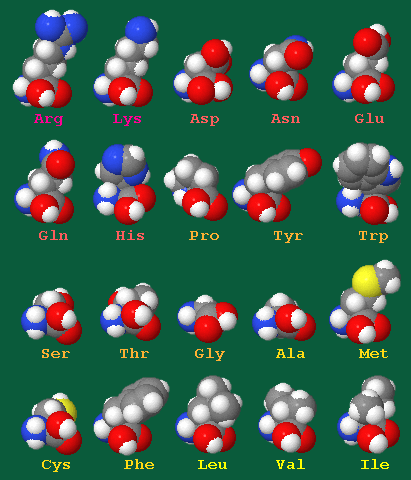
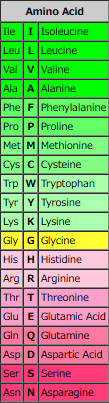
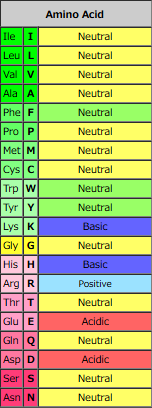
| Amino Acid | Molecular Formula | Hydropathy Index | log P | Acidic/Neutral/Basic | Polar/Hydrophobic | pI | ||
|---|---|---|---|---|---|---|---|---|
| Arg | R | Arginine | H2N(=NH)NHC3H6CCH(NH2)COOH | -4.5 | -4.20 | Acidic | Polar (Basic) | 10.76 |
| Lys | K | Lysine | H2N(CH2)4CH(NH2)COOH | -3.9 | -3.05 | Basic | Polar (Basic) | 9.74 |
| Asp | D | Aspartic Acid | HOOCCH2CH(NH2)COOH | -3.5 | -3.89 | Acidic | Polar (Acidic) | 2.77 |
| Asn | N | Asparagine | H2NCOCH2CH(NH2)COOH | -3.5 | -3.82 | Neutral | Polar (Neutral) | 5.41 |
| Glu | E | Glutamic Acid | HOOCCH2CH2CH(NH2)COOH | -3.5 | -3.69 | Acidic | Polar (Acidic) | 3.22 |
| Gln | Q | Glutamine | H2NCO-CH2CH2CH(NH2)COOH | -3.5 | -3.64 | Neutral | Polar (Neutral) | 5.65 |
| His | H | Histidine | H2NCH[CH2(C3H3N2)]COOH | -3.2 | -3.32 | Basic | Polar (Basic) | 7.59 |
| Pro | P | Proline | (C4H8N)-COOH | -1.6 | -2.54 | Neutral | Nonpolar (Hydrophobic) | 6.30 |
| Tyr | Y | Tyrosine | HO(C6H4)CH2CH(NH2)COOH | -1.3 | -2.26 | Neutral | Polar (Neutral) | 5.66 |
| Trp | W | Tryptophan | (C8H6N)-CH2CH(NH2)COOH | -0.9 | -1.05 | Neutral | Nonpolar (Hydrophobic) | 5.89 |
| Ser | S | Serine | HOCH2CH(NH2)COOH | -0.8 | -3.07 | Neutral | Polar (Neutral) | 5.68 |
| Thr | T | Threonine | CH3CH(OH)CH(NH2)COOH | -0.7 | -2.94 | Neutral | Polar (Neutral) | 6.16 |
| Gly | G | Glycine | H2NCH2COOH | -0.4 | -3.21 | Neutral | Nonpolar (Hydrophobic) | 5.97 |
| Ala | A | Alanine | CH3CH(NH2)COOH | 1.8 | -2.85 | Neutral | Nonpolar (Hydrophobic) | 6.00 |
| Met | M | Methionine | CH3SCH2CH2CH(NH2)COOH | 1.9 | -1.87 | Neutral | Nonpolar (Hydrophobic) | 5.74 |
| Cys | C | Cysteine | HSCH2CH(NH2)COOH | 2.5 | -2.49 | Neutral | Polar (Neutral) | 5.07 |
| Phe | F | Phenylalanine | (C6H5)CH2CH(NH2)COOH | 2.8 | -1.38 | Neutral | Nonpolar (Hydrophobic) | 5.48 |
| Leu | L | Leucine | (CH3)2CHCH2CH(NH2)COOH | 3.8 | -1.52 | Neutral | Nonpolar (Hydrophobic) | 5.98 |
| Val | V | Valine | (CH3)2CHCH(NH2)COOH | 4.2 | -2.26 | Neutral | Nonpolar (Hydrophobic) | 5.96 |
| Ile | I | Isoleucine | C2H5CH(CH3)CH(NH2)COOH | 4.5 | -1.70 | Neutral | Nonpolar (Hydrophobic) | 6.02 |
# log P; On-Line Log P Calculation
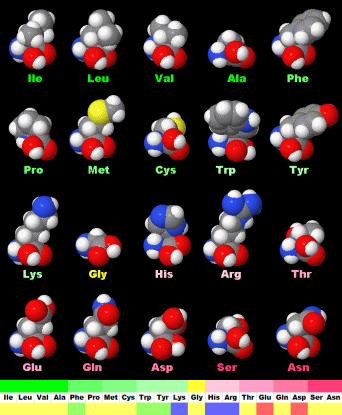
| Amino Acid | hydropathy index |
Side-chain R | log P | Acidic/Neutral/Basic | pI | ||||
|---|---|---|---|---|---|---|---|---|---|
| O | I | I/O * | |||||||
| Ile | I | Isoleucine | 4.5 | 80 | 0 | 0 | -1.70 | Neutral | 6.02 |
| Leu | L | Leucine | 3.8 | 70 | 0 | 0 | -1.52 | Neutral | 5.98 |
| Val | V | Valine | 4.2 | 50 | 0 | 0 | -2.26 | Neutral | 5.96 |
| Ala | A | Alanine | 1.8 | 20 | 0 | 0 | -2.85 | Neutral | 6.00 |
| Phe | F | Phenylalanine | 2.8 | 140 | 15 | 0.107 | -1.38 | Neutral | 5.48 |
| Pro | P | Proline | -1.6 | 60 | 10 | 0.167 | -2.54 | Neutral | 6.30 |
| Met | M | Methionine | 1.9 | 100 | 20 | 0.200 | -1.87 | Neutral | 5.74 |
| Cys | C | Cysteine | 2.5 | 60 | 20 | 0.333 | -2.49 | Neutral | 5.07 |
| Trp | W | Tryptophan | -0.9 | 180 | 130 | 0.722 | -1.05 | Neutral | 5.89 |
| Tyr | Y | Tyrosine | -1.3 | 140 | 115 | 0.821 | -2.26 | Neutral | 5.66 |
| Lys | K | Lysine | -3.9 | 80 | 70 | 0.875 | -3.05 | Basic | 9.74 |
| Gly | G | Glycine | -0.4 | 0 | 0 | - | -3.21 | Neutral | 5.97 |
| His | H | Histidine | -3.2 | 80 | 152 | 1.900 | -3.32 | Basic | 7.59 |
| Arg | R | Arginine | -4.5 | 80 | 190 | 2.375 | -4.20 | Positive | 10.76 |
| Thr | T | Threonine | -0.7 | 40 | 100 | 2.500 | -2.94 | Neutral | 6.16 |
| Glu | E | Glutamic Acid | -3.5 | 60 | 150 | 2.500 | -3.69 | Acidic | 3.22 |
| Gln | Q | Glutamine | -3.5 | 60 | 200 | 3.333 | -3.64 | Neutral | 5.65 |
| Asp | D | Aspartic Acid | -3.5 | 40 | 150 | 3.750 | -3.89 | Acidic | 2.77 |
| Ser | S | Serine | -0.8 | 20 | 100 | 5.000 | -3.07 | Neutral | 5.68 |
| Asn | N | Asparagine | -3.5 | 40 | 200 | 5.000 | -3.82 | Neutral | 5.41 |
 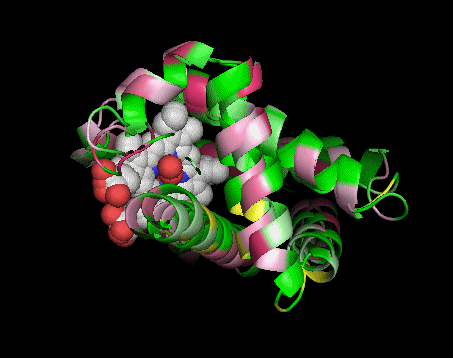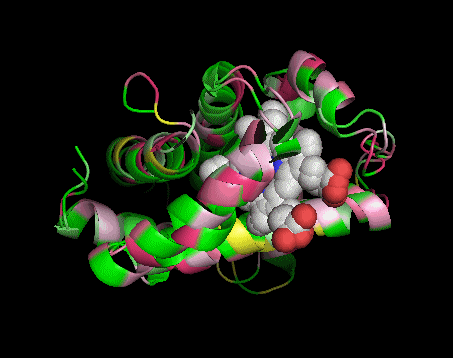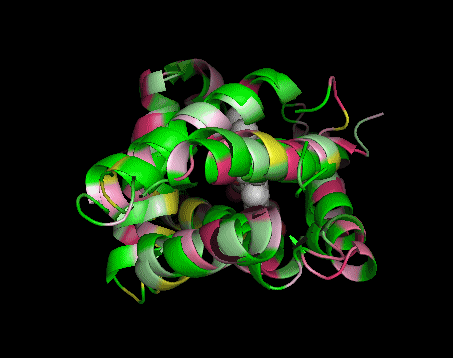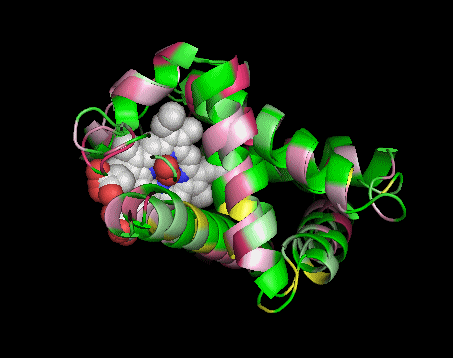 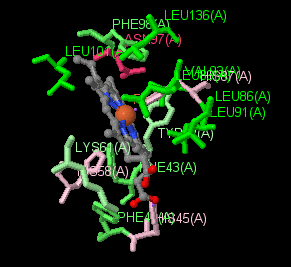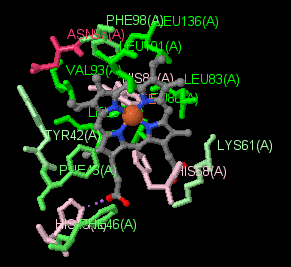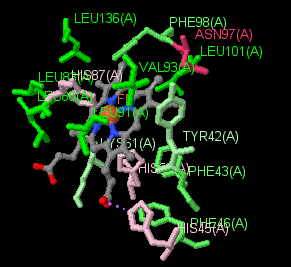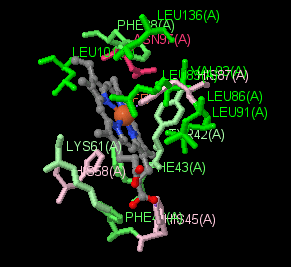
|
  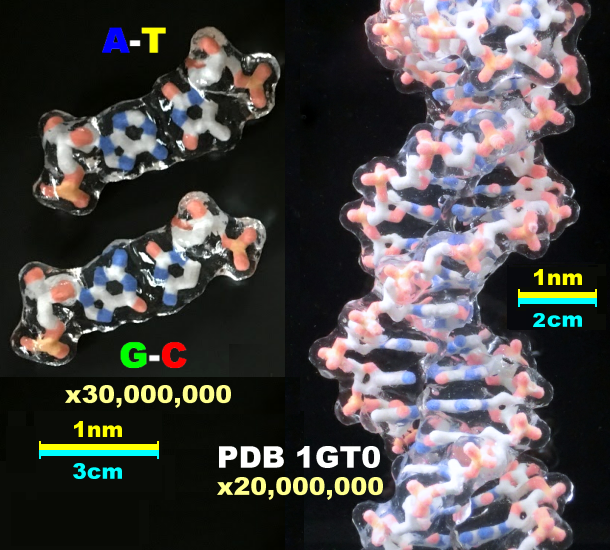
|

Booth 327 - Science Agora 2024
Conformational Preferences of the Amino Acids
Williams, R.W. et al. : Biochim. Biophys. Acta , 916, 200-204 (1987).
"From Sequence to Structure" [PDF], p.18
α-helix
β-strand
Glu
1.59
0.52
Ala
1.41
0.72
Leu
1.34
1.22
Met
1.30
1.14
Gln
1.27
0.98
Lys
1.23
0.69
Arg
1.21
0.84
His
1.05
0.80
Val
0.90
1.87
Ile
1.09
1.67
Tyr
0.74
1.45
Cys
0.66
1.40
Trp
1.02
1.35
Phe
1.16
1.33
Thr
0.76
1.17
Gly
0.43
0.58
Asn
0.76
0.48
Pro
0.34
0.31
Ser
0.57
0.96
Asp
0.99
0.39